Getting started with FISPACT-II
Thomas Stainer
23rd November 2020
Useful links
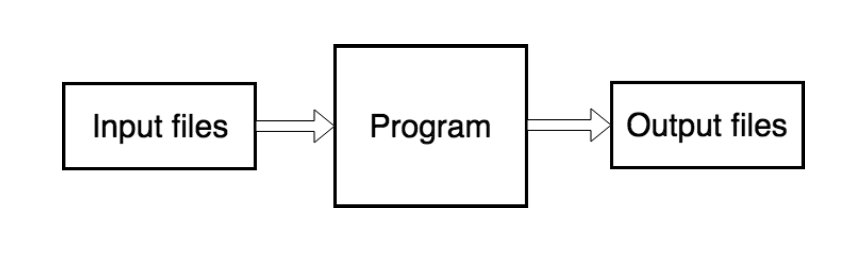
What is FISPACT-II?
- Command line tool only
- Works on Windows, Linux and Mac
- Single binary executable program
- File driven program (via input files)
- Produces output files (text, TAB and JSON formats)
- Human readable outputs with more structured outputs for parsing
How to run FISPACT-II
- No GUI (yet)
- Use via terminal (Linux) or cmd (Windows)
- Powershell or Cygwin can be used too!
Be aware
- Windows uses .exe extension (not the case for Mac and Linux)
- Windows uses \ not / (Linux/Mac)
- Important for files file (more on that later). Linux style paths can be used on Windows
- Input and output files are independent of operating system
Running FISPACT-II
- Requires input files as arguments
- Will not run without an input
Mac/Linux
[tom@mac:~/Dev/fispact]$ /path/to/fispact # gives an errorWindows
C:\Dev\fispact> C:\path\to\fispact.exe # gives an errorRunning FISPACT-II correctly
We need an input file myinput.i and files file myfiles (always!)
[tom@mac:~/Dev/fispact]$ /path/to/fispact myinput myfiles
Running....
Note that .i is needed for the input file name extension but is omitted from cmd line
[tom@mac:~/Dev/fispact]$ ls
myinput.i myfiles
[tom@mac:~/Dev/fispact]$ /path/to/fispact myinput myfiles
Running FISPACT-II correctly
- First argument is the input file myinput.i
- Second argument is the files file filesfile (optional)
- Input file must have .i extension
- Files file can have extension (if you want) but normally doesn't
- Files file argument can be omitted iff it is named 'files', 'Files', or 'FILES'
Note that .i is needed for the input file name extension but is omitted from cmd line
[tom@mac:~/Dev/fispact]$ ls
myinput.i myfiles FILES
# uses myfiles file
[tom@mac:~/Dev/fispact]$ /path/to/fispact myinput myfiles
# uses FILES file
[tom@mac:~/Dev/fispact]$ /path/to/fispact myinput
Creating an alias for FISPACT-II
Instead of typing
/path/to/fispactEasier to make an alias
Mac/Linux
[tom@mac:~/Dev/fispact]$ export FISPACT=/path/to/fispact[tom@mac:~/Dev/fispact]$ $FISPACT myinput myfilesWindows
C:\Dev\fispact> set FISPACT=C:\path\to\fispact.exeC:\Dev\fispact> %FISPACT% myinput myfilesPoll: FISPACT-II experience
Click here to take the pollFiles file
- We will see later how to make these files from scratch
- But first let's learn more about the files file
- Why do we need it?
- What does it do?
- What do we need to put in it?
Files file
The files file declares the nuclear data libraries to be used in the fispact run*
(*mainly -it also tells FISPACT-II the names and locations of files to read and write)
FISPACT-II is useless without nuclear data!
Files file
- A text file
- With a list of keys and values
- Typically named 'files', 'Files' or 'FILES'
- But can be named anything e.g. files.tendl2015.run1
- Order of paths does not matter
- Use linux style '/' for paths
- Don’t use tab to indent – causes parsing problems!
Files file
Nuclear data paths...
# index of nuclides to be included
ind_nuc /home/F-II/nuclear_data/TENDL2015data/tendl15_decay12_index
# Library cross section data
xs_endf /home/F-II/nuclear_data/TENDL2015data/tal2015-n/gxs-709
# Library decay data
dk_endf /home/F-II/nuclear_data/decay/decay_2012Files file
Other data paths...
# fluxes file to be read in
fluxes fluxes
# collapsed cross section data (in and out)
collapxi COLLAPX
collapxo COLLAPX
# condensed decay and fission
arrayx ARRAYXFiles file
- Normally use about 12 - 16 key-value pairs needed
- Over 40 possible options
- See advanced usage for details on the others
- In most examples here, we will use no more than 16.
Nuclear data
- FISPACT-II can process a lot of different nuclear data libraries
- Can process both ENDF-6 format and EAF format libraries
- A large number are shipped with version 4.0 already
- FISPACT-II uses group wise data, not point wise
Disclamer: FISPACT-II does not hold responsibility for nuclear data errors/issues, it is simply a consumer of nuclear data
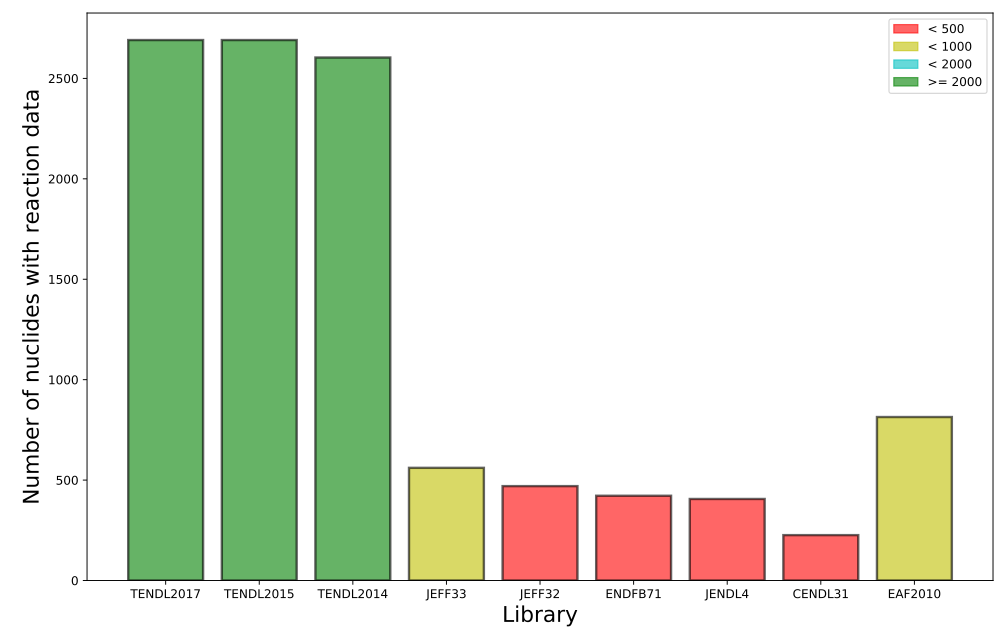
Nuclear data: quick compare
Reaction data
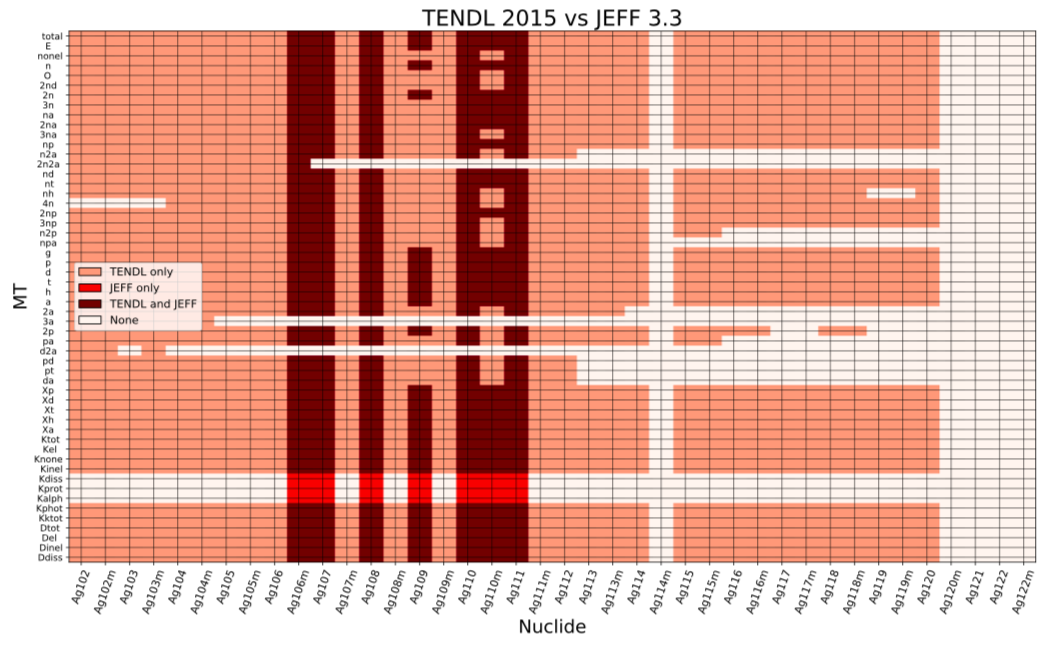
Nuclear data: in depth compare
Reaction data for Silver
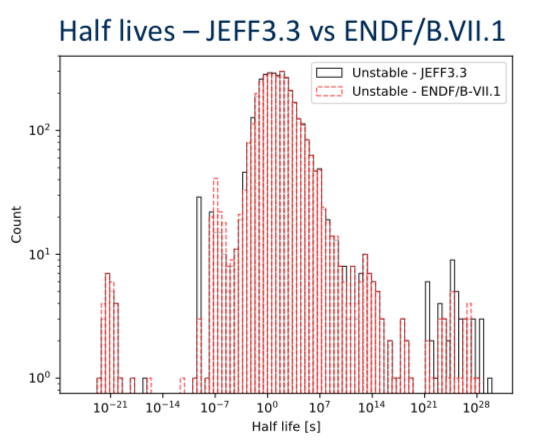
Decay data is different too
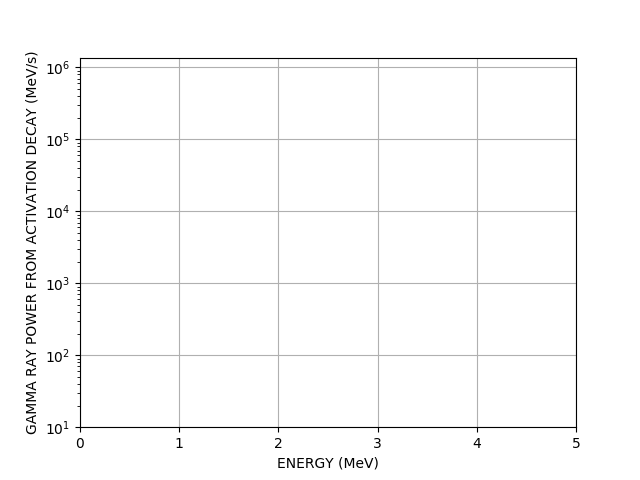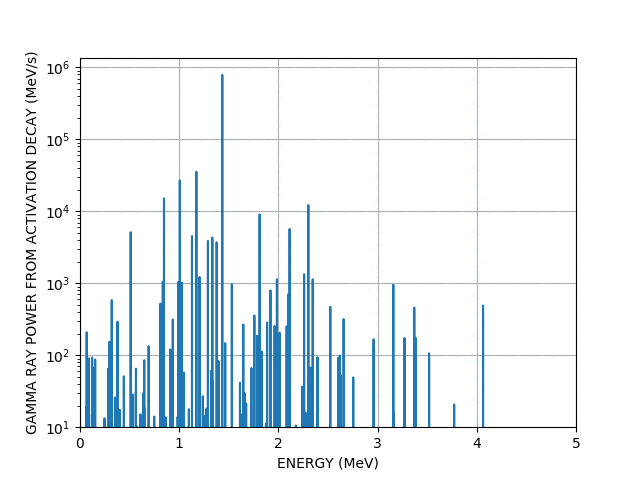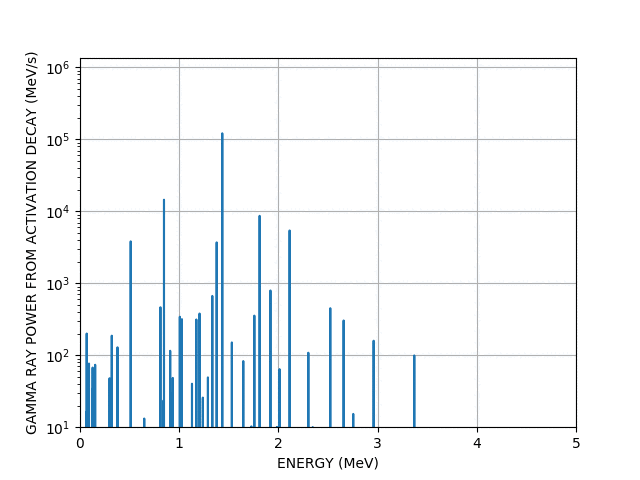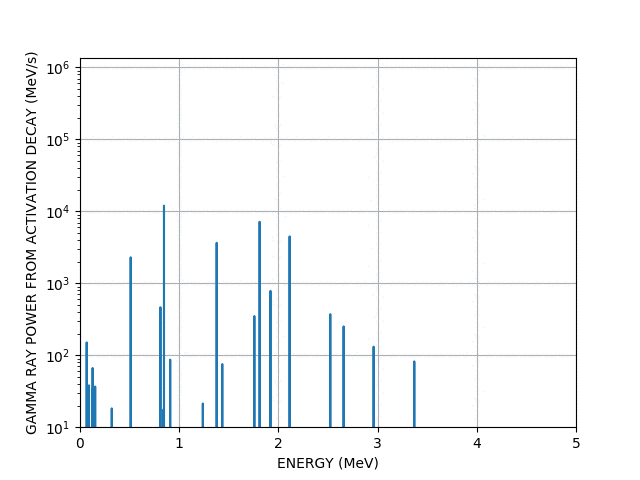
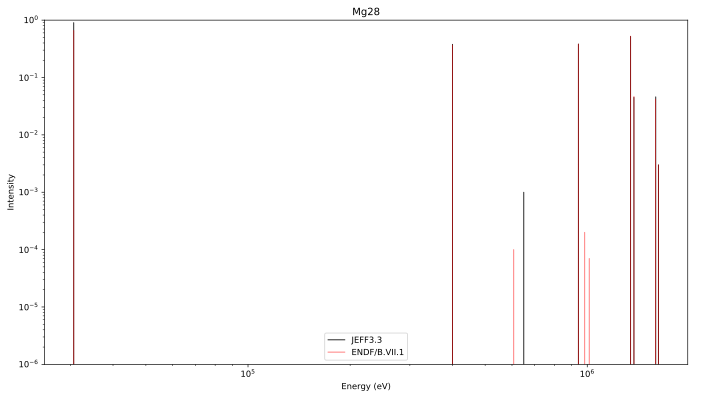
Gamma lines
JEFF3.3 vs ENDFB VII

A final note on nuclear data
TENDL recommended
- FISPACT-II development has run in parallel with that of the TALYS-based Evaluated Nuclear Data Library – TENDL*
- It is the largest and most complete nuclear data library!
- 2014, 2015 and 2017 included with FISPACT-II
- 2019 also now available
* A. J. Koning, D. Rochman, et al. - https://tendl.web.psi.ch/tendl_2017/tendl2017.html
3850+ nuclides
Full decay_2012 library

Compressing nuclear data
- Large nuclear data libraries (such as TENDL) can take a while to read
- Sometimes up to 2 minutes for TENDL 2017
- This can be improved by up to 60% using the compress_xs_endf tool
- Separate tool shipped with FISPACT-II
- Turns nuclear data text files into binary equivalent
- Note: they are OS dependant
- Use GETXS -1 for binary files
Core nuclear data
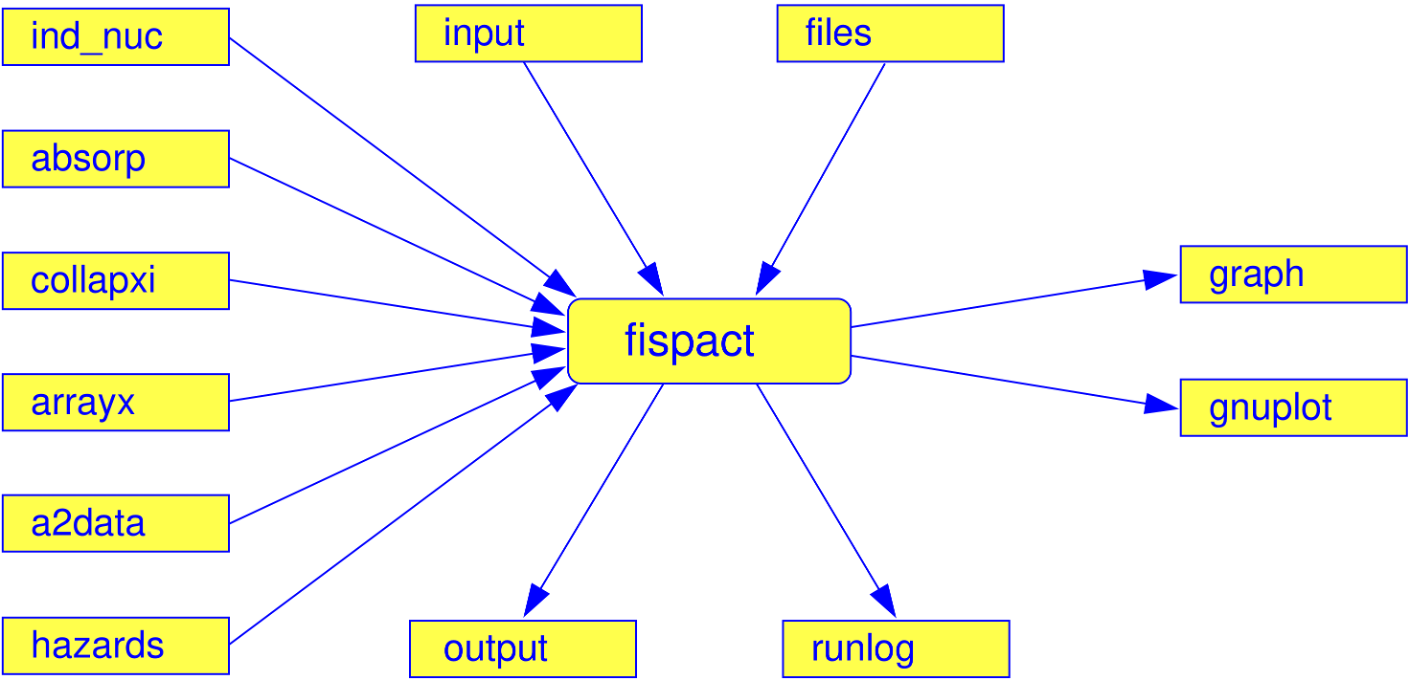
- ind_nuc = Index file – tells what nuclides to include in the simulation
- xs_endf = Cross section data directory (ENDF-6 format)
- xs_endfb = Cross section data directory (compressed binary format)
- dk_endf = Decay data directory (ENDF-6 format)
- a2data = file path of transportation values of radioactive material
- clear = file path of clearance values of radioactive material
- hazards = file path of ingestion and inhalation values for radionuclides
- absorp = file path of air attenuation data for nuclides
- fy_endf = neutron fission yield data directory
- sf_endf = spontaneous fission yield data directory
- prob_tab = directory path to probability table data for self shielding
Note: For EAF keywords can differ. For example, xs_endf -> crossec
Other important data
As mentioned before, files file has other data
- fluxes = Defines your flux (incident particle energy spectrum) - always needed for irradiation simulations
- collapxi = if using a pre collapsed binary file, use this instead (more details later)
- collapxo = pre collapsed binary file output path
- arrayx = binary file for decay data output path
Fluxes file
Defines your incident particle energy spectrum
- If you want to do irradiation then a fluxes file is needed
- We have to tell FISPACT-II in the files file about the fluxes file
- The file contains the group wise data as a list of values
- Note: Data must be specified in descending order (high to low energy)!
- Group structure must match that of nuclear data
- e.g. If using TENDL2017 then you will need your spectra defined in 709 group.
- Group convert (GRPCONVERT) can change group structure (more on this later)
Fluxes file


group 616
group 709
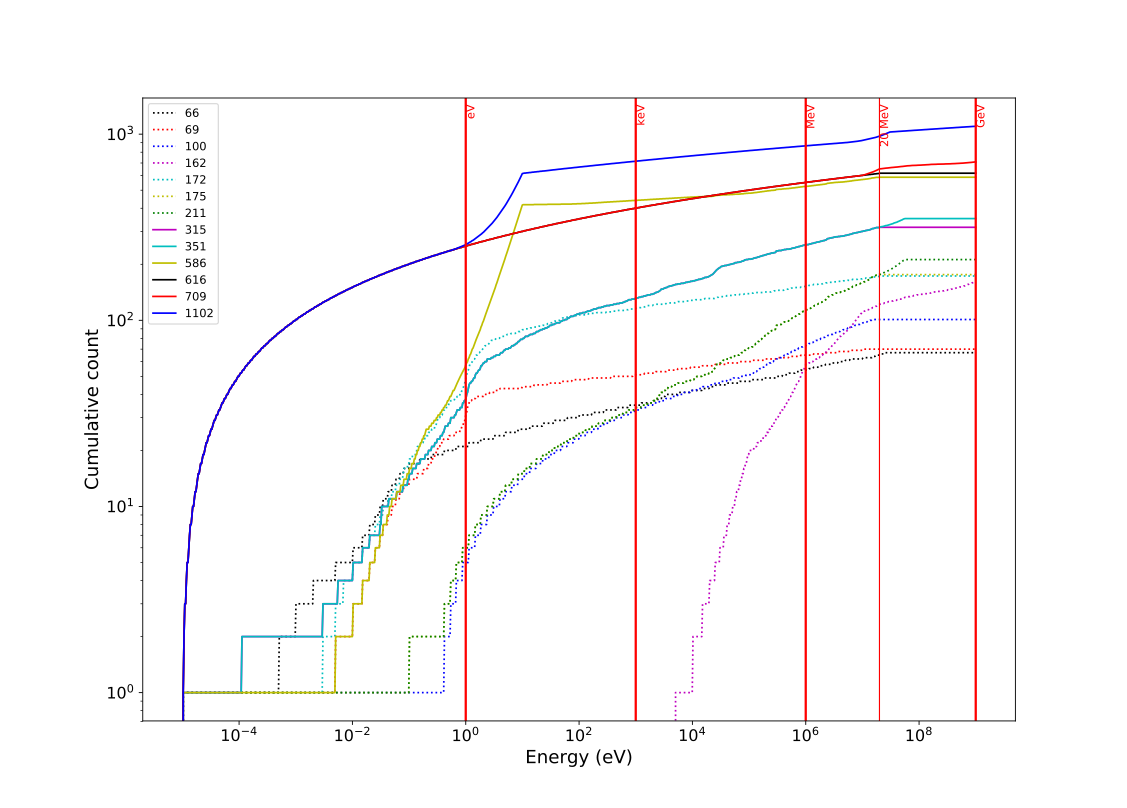
Group structures
Different group structures cover different energy regimes
Some libraries are provided in a given energy range best suited for certain applications
709 group covers up to 1 GeV – most libraries shipped in this structure
Input file
The input file governs everything for the run
- Material definition
- Irradiation schedule
- Log options
- Uncertainties
- Solver settings
- Self shielding
- Pathways
- Output types – TAB, JSON, graph files
Input file
- A text file
- List of keywords and arguments
- No default name, unlike the files file
- Input file must have .i extension
- Order of paths does matter (unlike files file)
- Allows comments with ‘<< comment here >>’
- Note: There is a limit of 120 characters per line!
- 92 keywords in all!
Input file
3 sections to the input file
- Control - 32 possible keywords
- Initialisation - 64 possible keywords
- Inventory - 41 possible keywords
32 + 64 + 41 ≠ 92
Many keywords can be used in each input section, but some can only be used in one section!
Full list: https://fispact.ukaea.uk/wiki/FISPACT-II_keywords
Simple example
<< CLOBBER tells fispact to overwrite >>
<< existing files of same name >>
CLOBBER
<< GETXS 1 tells fispact to get the cross section>>
<< data and do collapse >>
GETXS 1 709
<< GETDECAY 1 tells fispact to get the decay>>
<< data and do condense >>
GETDECAY 1
<< FISPACT signals end of control section >>
FISPACT
* example 1
<< END is required to signal end of input >>
END
* end of example
Control only - no Initialisation or inventory sections here
Collapse
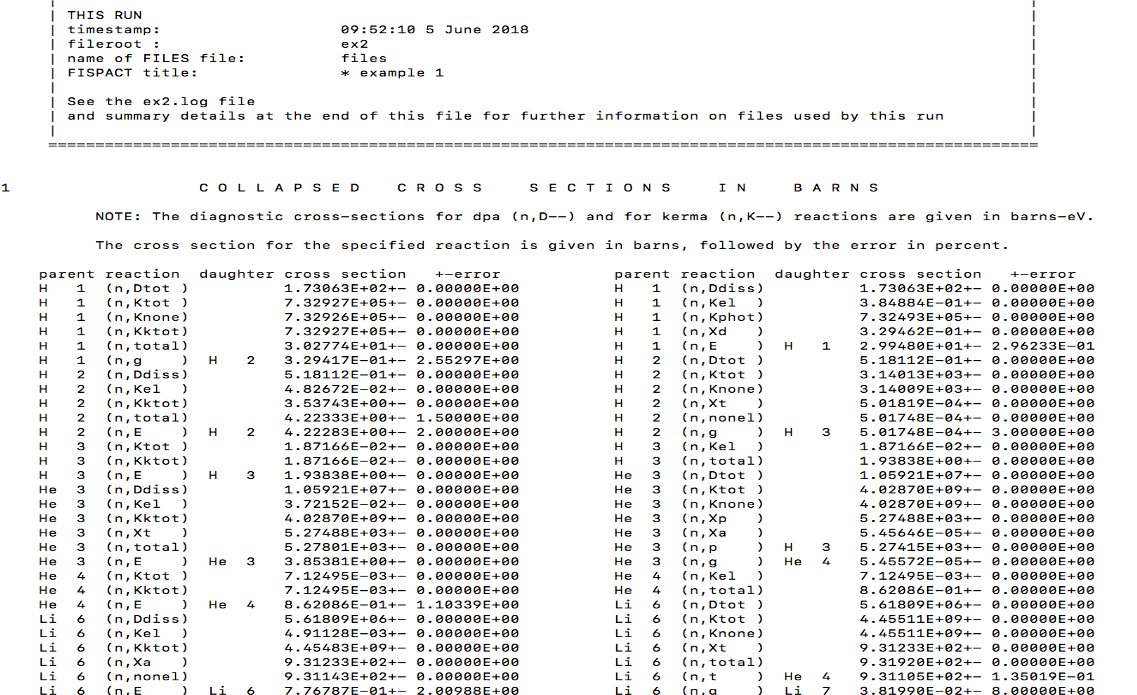
The nuclear data collapse with GETXS takes the extensive reaction cross section data and probability tables for resonance self-shielding and folds (collapses) them with irradiation spectrum in fluxes. Producing a single scalar value (1-group).
Collapse
Produces a binary COLLAPX file of 1-group data with uncertainties which is given by the collapxo file in the files file
\[\begin{aligned} \bar{\sigma} & = \frac{\sum^{N}_{i}\sigma_{i}\phi_{i}}{\sum^{N}_{i}\phi_{i}} \end{aligned} \]Exercise: printlib
Printlib 4 prints all collapsed cross sections to disk
The cross sections are given for each reaction
See the exercise on this.
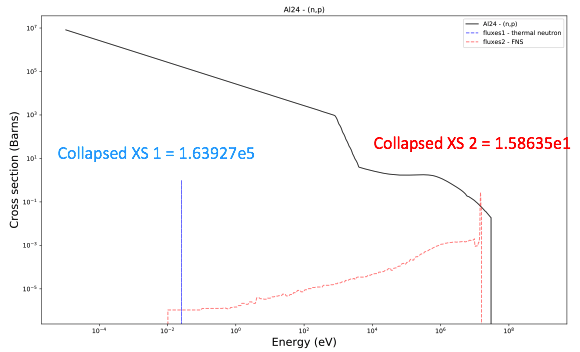
Exercise: printlib
Can actually calculate this by hand
Getting the group wise XS data can be done using another tool with FISPACT-II extract_xs_endf
Collapse and condense
Common practice is to separate out collapse and condense.
collapse.i input file – GETXS 1
condese.i input file – GETDECAY 1
Computationally expensive to do these processes. If flux is not changing, then no need to redo collapse. We just did the collapse in the previous example.
[tom@mac:~/exercises/basic/inventory]$ $FISPACT collapse
[tom@mac:~/exercises/basic/inventory]$ $FISPACT condense
[tom@mac:~/exercises/basic/inventory]$ $FISPACT input1
[tom@mac:~/exercises/basic/inventory]$ $FISPACT input2
Condense
The nuclear data condense via GETDECAY produces a binary ARRAYX file from the extensive decay and fission yield data
May also require an irradiation spectrum (fluxes) to calculate spectrum-dependent fission yields but is not normally the case
Collapse summary
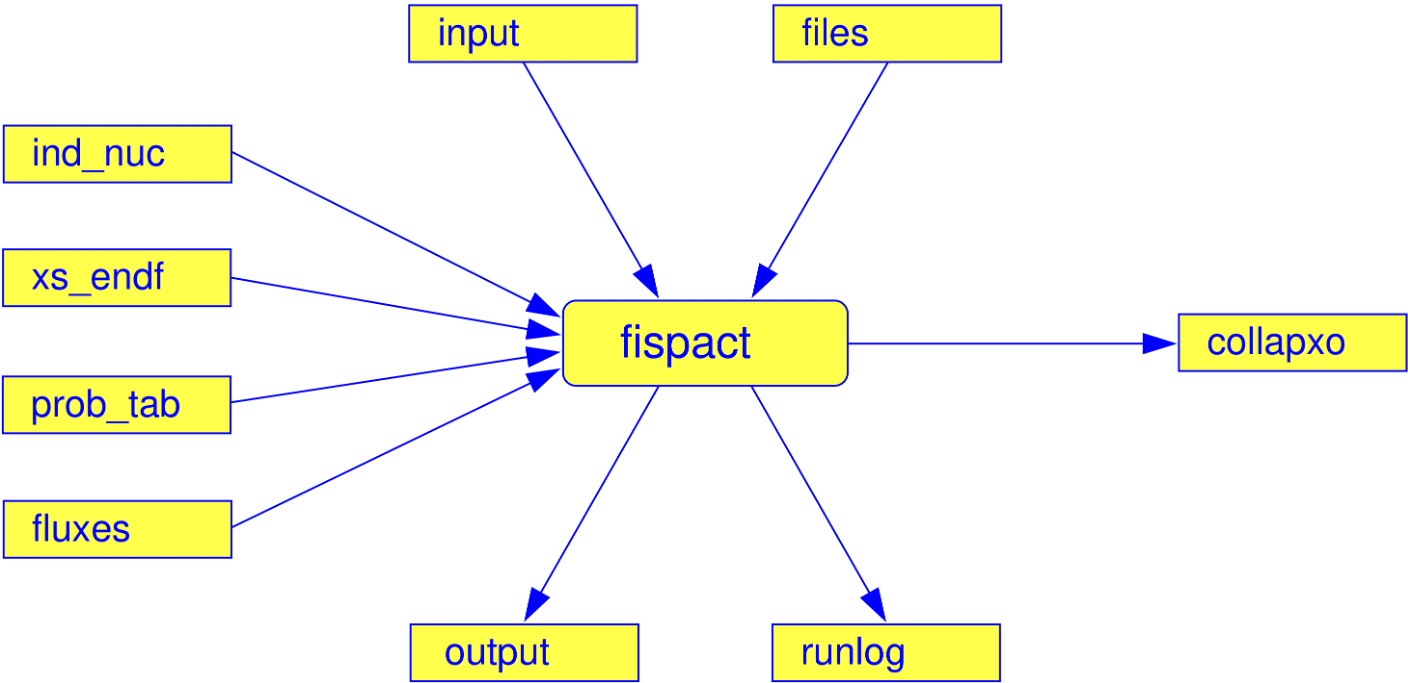
Condense summary

Inventory summary
Summary so far
- What FISPACT-II is
- Some of what FISPACT-II can do
- What files FISPACT-II needs
- Files file
- Fluxes file
- Input file
- How to run FISPACT-II
In the basic exercises we will use the fluxes and nuclear data to calculate all of the effective cross sections and prepare the decay data for time-dependent simulations.
However, all of this is not very interesting. No actual irradiation yet.
Worked example
FNS Inconel (from getting started)
Contains 4 input files
- collapse.i
- condense.i
- printlib.i
- inventory.i
First 3 have been covered, focus on the inventory run
Worked example - control
<< Overwrite any existing output file >>
CLOBBER
<< we also want a JSON output file >>
JSON
<< read in COLLAPX file>>
GETXS 0
<< read in ARRAYX file>>
GETDECAY 0
<< end of control section >>
FISPACT
* FNS 5 Minutes Inconel-600
Worked example - initial conditions I
<< Inconel 600 with density of 8.42 g/cc >>
DENSITY 8.42
<< 1 gram with 4 elements >>
MASS 1.0E-3 4
<< 75.82% Ni >>
NI 75.82
<< 15.97% Cr >>
CR 15.97
<< 7.82% Fe >>
FE 7.82
<< 0.39% Mn >>
MN 0.39Worked example - initial conditions II
<< ignore nuclides with inventory < 1000 atoms >>
MIND 1E3
<< create gnuplot data and file >>
GRAPH 1 2 1 3
<< pathways analysis >>
UNCERTAINTY 2
<< output half lives >>
HALF
<< output ingestion and inhalation >>
HAZARDSWorked example - irradiation phase
<< irradiate material at 1.116e10 n/cm2/s >>
FLUX 1.116E+10
<< initial inventory (t=0) >>
ATOMS
<< want output after 5 minutes irradiation >>
TIME 5.0 MINS
<< solves rate equation matrix and >>
<< outputs initial inventory at time=5 mins>>
ATOMS
Note: Without either ATOMS, STEP, or SPECTRUM, nothing will be calculated! For less verbose output, use STEP, instead of ATOMS.
Worked example - cooling phase
<< turn flux off - decay only >>
FLUX 0.
<< tells FISPACT-II we are cooling >>
ZERO
<< cooling time intervals >>
<< 38 seconds after irradiation ended >>
TIME 36 ATOMS
<< 15 seconds after previous time >>
TIME 15 ATOMS
<< 16 seconds after previous time >>
TIME 16 ATOMS
...
<< end of input file >>
END
* ENDNote: Times are intervals. It is time difference not cumulative time
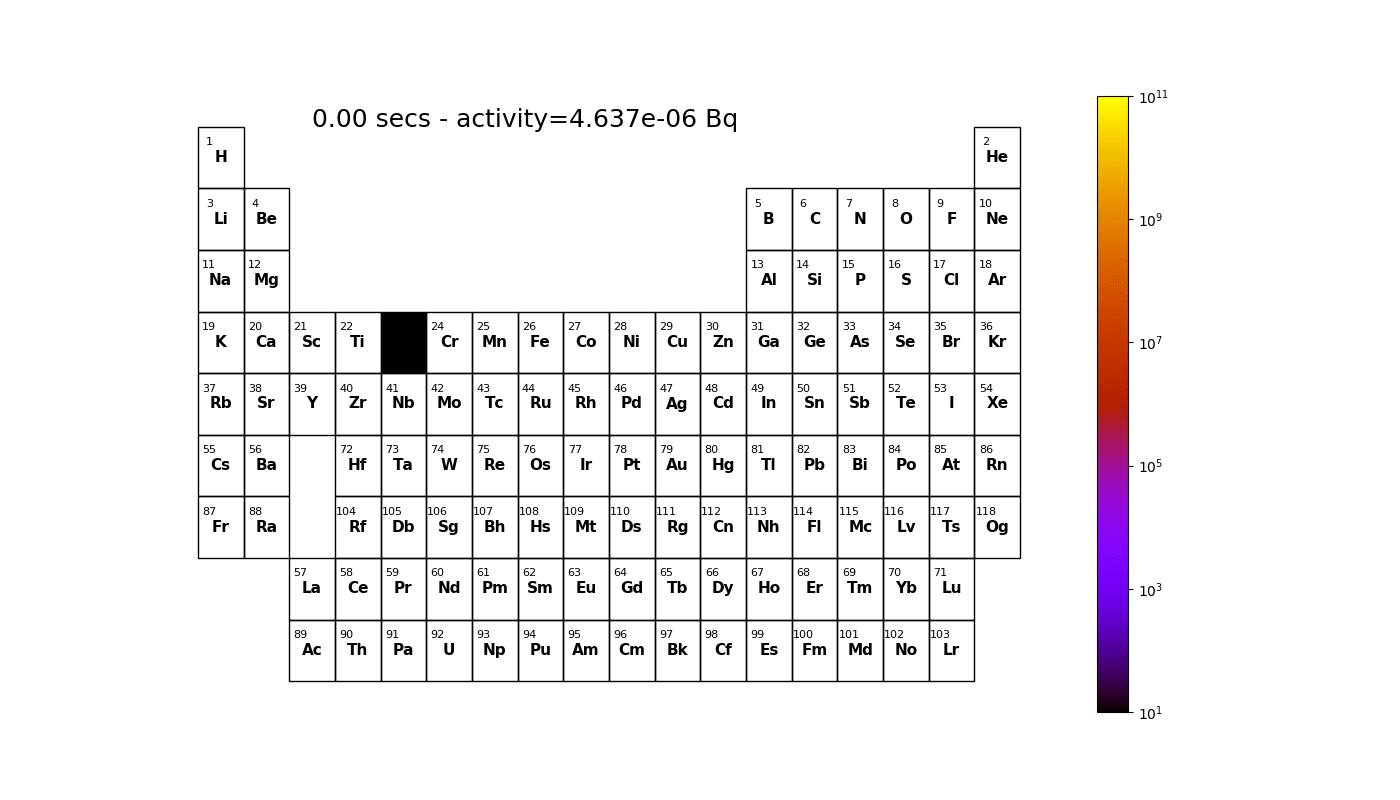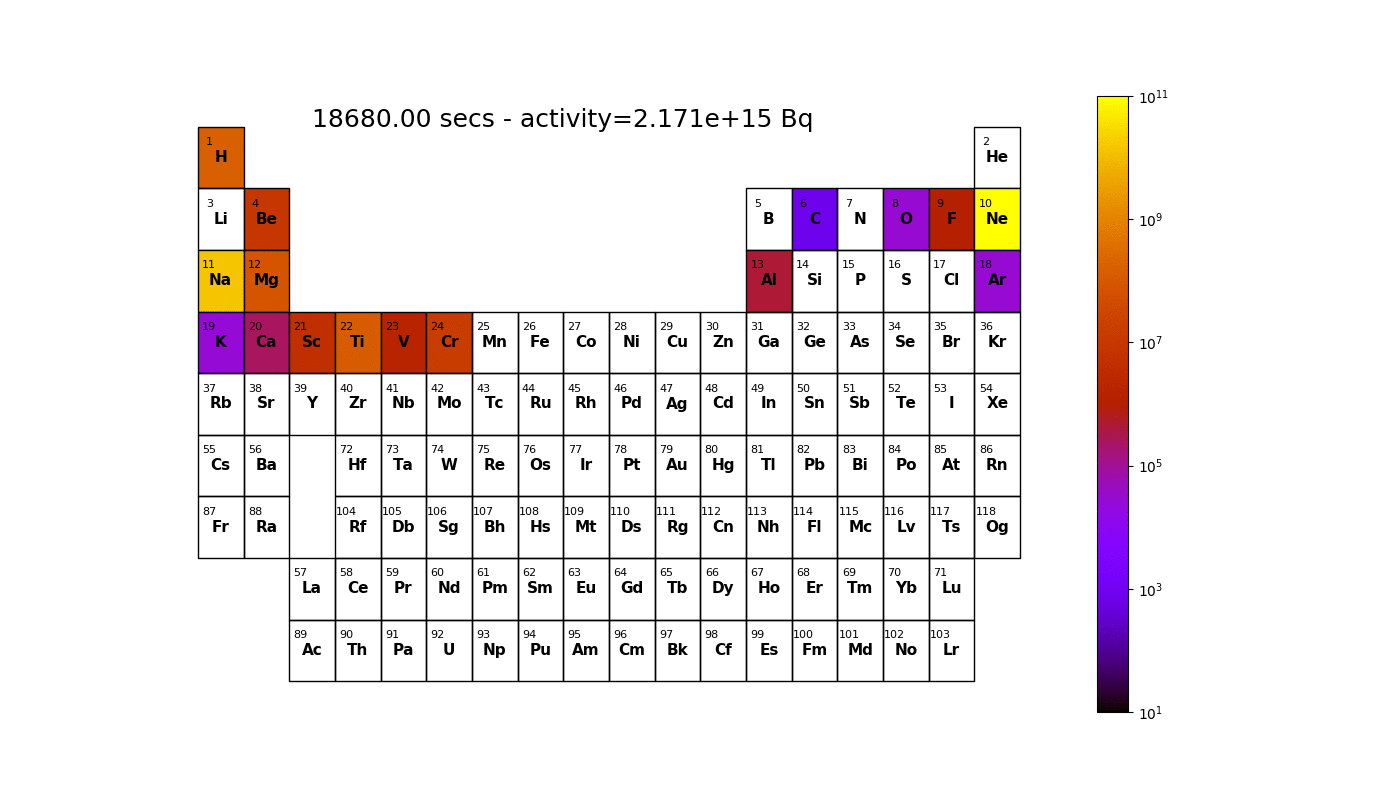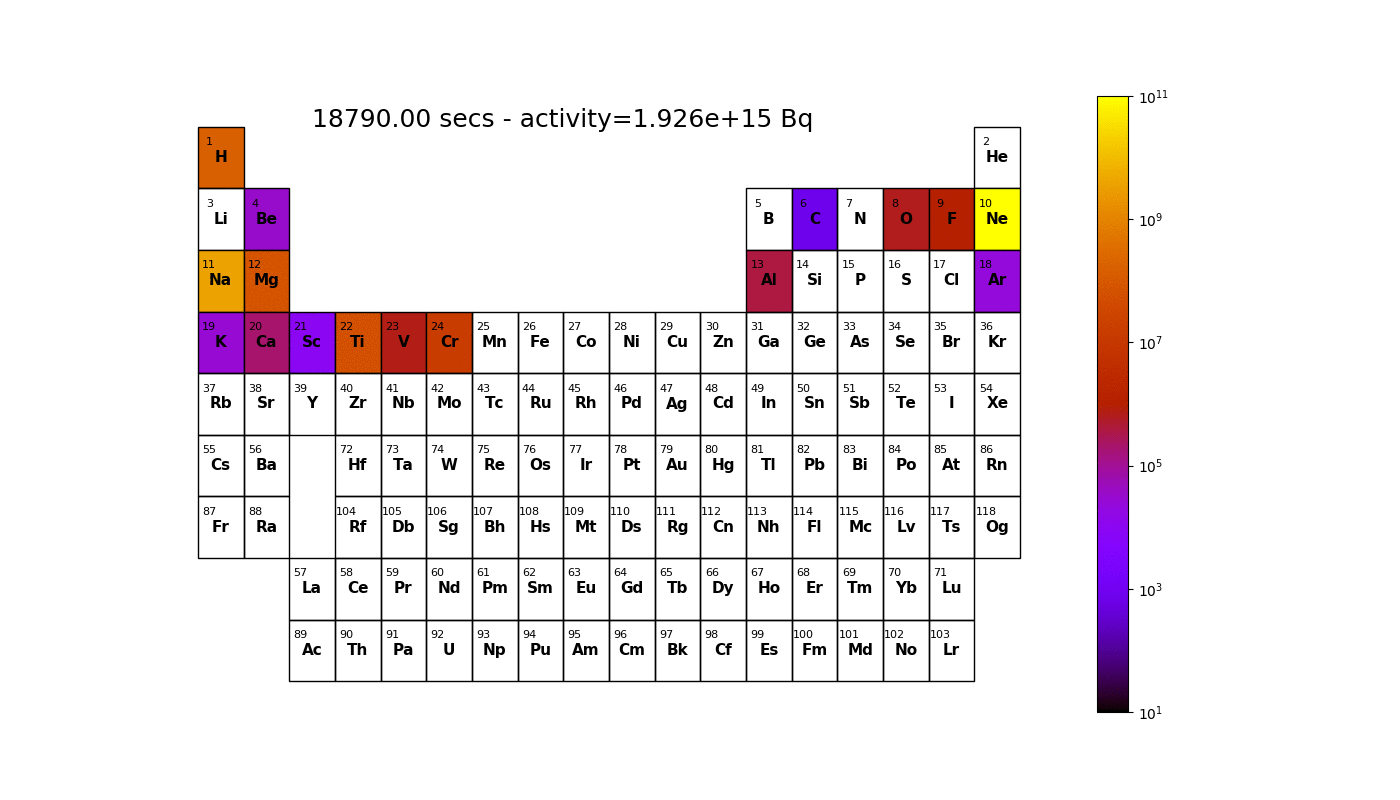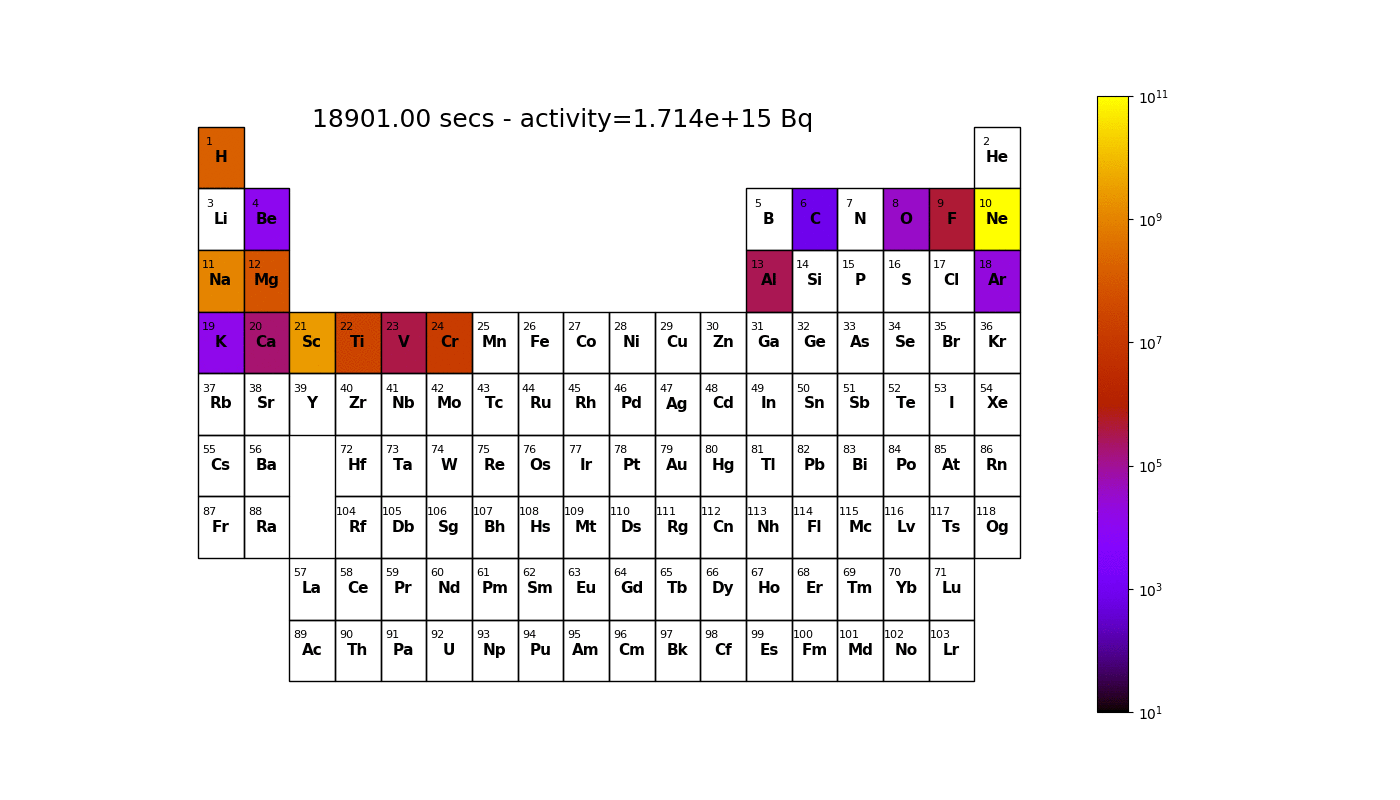
Worked example - graph outputs

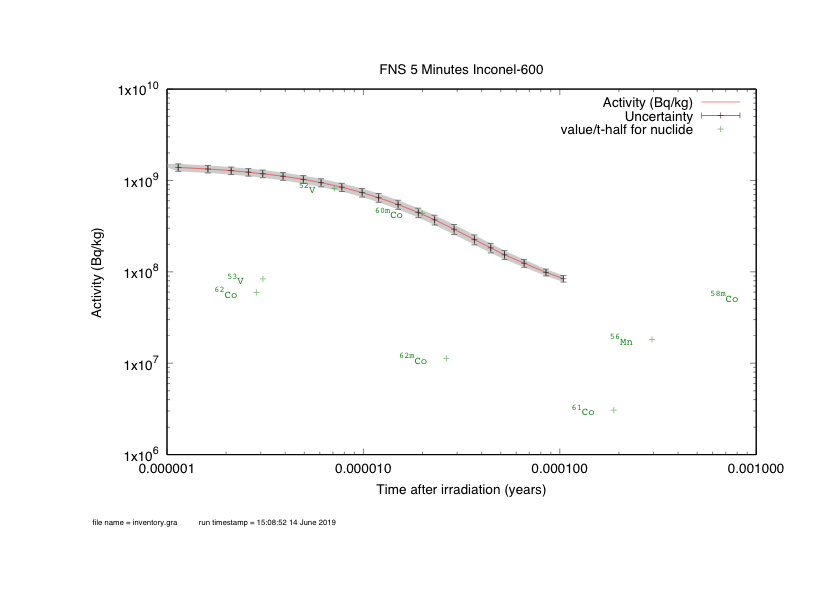
Worked example summary
Able to irradiate a material and generate graphs
But what if we want custom graphs?
- Generated graphs and plots are good, but limited
- We can look and copy values from the output file, but difficult for big simulations
- Output file can be tricky to parse
- Other output files – TAB and JSON to help file reading issues
- FISPACT-II official parser - pypact
Pypact
Can make parsing legacy FISPACT-II files easy - python3 package